Genotyping method and frequency of single nucleotide polymorphism rs12970134 near melanocortin-4 receptor genotypes in hanoi preschool chidren population
Melanocortin-4 receptor is part of the central melanocortinergic system and plays critical roles in central
regulation of food intake, energy homeostasis and body weight, so that this gene is related to obesity
and insulin resistance including single nucleotide polymorphism rs12970134. Thus, this study aimed to
find out a protocol for genotyping rs12970134 near Melanocortin-4 receptor by AS-PCR method,
analyzing the genotype and allele ratios of this single nucleotide polymorphism in 200 3-5 years old
children in Hanoi, Vietnam (50% boys). This protocol used the forward primers including 5’-
tcttaccaaacaaagcatgtg-3’ to detect allele G, and 5’-tcttaccaaacaaagcatgta-3’ to detect allele A; and the
reverse primer 5’-gtcattcccactaccacctg-3’. The optimized PCR protocol was that 94oC for 3 min and 34
cycles of denaturation at 94oC for 30 sec, primer annealing at 54oC for 40 sec, primer extension at 72oC
for 30 sec, final extension at 72oC for 8 min, stopped by chilling at 4oC. The 208 bp PCR products were
detected on Redsafe-stained 2.5% agarose gel. The results were verified by using the sequencing
method. In the entire samples, the GG genotype was the largest (57.5%), and the AA genotype was the
lowest (4%). The frequencies of the G and A alleles were 0.77 and 0.23, respectively.

Trang 1

Trang 2

Trang 3

Trang 4
Trang 5

Trang 6

Trang 7
Tóm tắt nội dung tài liệu: Genotyping method and frequency of single nucleotide polymorphism rs12970134 near melanocortin-4 receptor genotypes in hanoi preschool chidren population

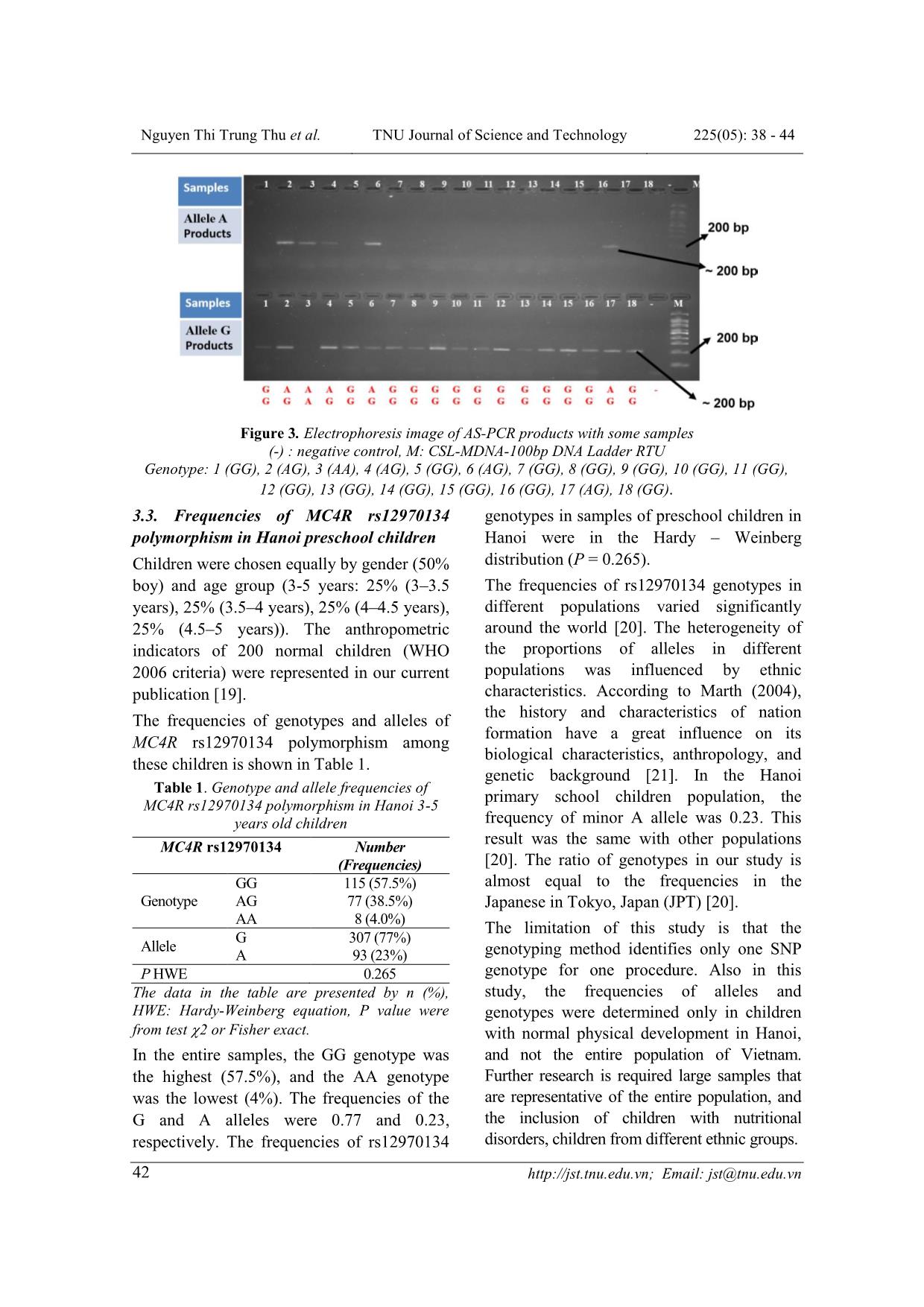
ISSN: 1859-2171 e-ISSN: 2615-9562 TNU Journal of Science and Technology 225(05): 38 - 44 38 Email: jst@tnu.edu.vn GENOTYPING METHOD AND FREQUENCY OF SINGLE NUCLEOTIDE POLYMORPHISM RS12970134 NEAR MELANOCORTIN-4 RECEPTOR GENOTYPES IN HANOI PRESCHOOL CHIDREN POPULATION Nguyen Thi Trung Thu, Le Thi Tuyet * Hanoi National University of Education ABSTRACT Melanocortin-4 receptor is part of the central melanocortinergic system and plays critical roles in central regulation of food intake, energy homeostasis and body weight, so that this gene is related to obesity and insulin resistance including single nucleotide polymorphism rs12970134. Thus, this study aimed to find out a protocol for genotyping rs12970134 near Melanocortin-4 receptor by AS-PCR method, analyzing the genotype and allele ratios of this single nucleotide polymorphism in 200 3-5 years old children in Hanoi, Vietnam (50% boys). This protocol used the forward primers including 5’- tcttaccaaacaaagcatgtg-3’ to detect allele G, and 5’-tcttaccaaacaaagcatgta-3’ to detect allele A; and the reverse primer 5’-gtcattcccactaccacctg-3’. The optimized PCR protocol was that 94oC for 3 min and 34 cycles of denaturation at 94 o C for 30 sec, primer annealing at 54 o C for 40 sec, primer extension at 72 o C for 30 sec, final extension at 72 o C for 8 min, stopped by chilling at 4 o C. The 208 bp PCR products were detected on Redsafe-stained 2.5% agarose gel. The results were verified by using the sequencing method. In the entire samples, the GG genotype was the largest (57.5%), and the AA genotype was the lowest (4%). The frequencies of the G and A alleles were 0.77 and 0.23, respectively. Key words: Melanocortin-4 receptor; single nucleotide polymorphism; rs12970134; AS-PCR method; preschool children. Received: 09/02/2020; Revised: 27/4/2020; Published: 28/4/2020 PHƯƠNG PHÁP PHÂN TÍCH KIỂU GEN VÀ TẦN SỐ ALEN CỦA ĐA HÌNH NUCLEOTIDE ĐƠN RS12970134 GẦN THỤ THỂ MELANOCORTIN-4 Ở TRẺ MẦM NON TẠI HÀ NỘI Nguyễn Thị Trung Thu, Lê Thị Tuyết* Trường Đại học Sư phạm Hà Nội TÓM TẮT Thụ thể Melanocortin-4 có liên quan đến bệnh béo phì, kháng insulin do đóng vai trò quan trọng trong việc điều hòa lượng thức ăn ăn vào, cân bằng nội môi và khối lượng cơ thể. Mục tiêu của nghiên cứu này là xây dựng được phương pháp AS-PCR phân tích kiểu gen của đa hình nucleotide đơn rs12970134 gần thụ thể Melanocortin-4 và xác định tỉ lệ alen của đa hình nucleotide đơn này ở trẻ em 3-5 tuổi tại Hà Nội. Nghiên cứu đã thiết kế được các đoạn mồi để xác định alen của đa hình nucleotide đơn rs12970314 gồm mồi xuôi phát hiện alen G: 5'-tcttaccaaacaaagcatgtg-3', phát hiện alen A: 5'-tcttaccaaacaaagcatgta-3'; và mồi ngược: 5'-gtcattcccactaccacctg-3'. Chu trình nhiệt của phản ứng PCR được tối ưu hóa là: 94oC (3 phút) và 34 chu kỳ: biến tính ở 94oC (30 giây), gắn mồi ở 54oC (40 giây), kéo dài ở 72oC (30 giây), bước kéo dài cuối ở 72oC (8 phút), kết thúc ở 4oC. Sản phẩm PCR 208 bp được phát hiện trên gel agarose 2,5% nhuộm redsafe. Kiểu gen được kiểm tra bằng phương pháp giải trình tự gen. Trong toàn bộ mẫu, tỉ lệ kiểu gen GG là lớn nhất (57,5%) và AA là thấp nhất (4%). Tần số của các alen G và A lần lượt là 0,77 và 0,23. Từ khóa: Thụ thể Melanocortin-4; đa hình nucleotide đơn; rs12970134; phương pháp AS-PCR; trẻ mầm non. Ngày nhận bài: 09/02/2020; Ngày hoàn thiện: 27/4/2020; Ngày đăng: 28/4/2020 * Corresponding author. Email: lttuyet@gmail.com DOI: https://doi.org/10.34238/tnu-jst.2020.05.2604 Nguyen Thi Trung Thu et al. TNU Journal of Science and Technology 225(05): 38 - 44 Email: jst@tnu.edu.vn 39 1. Introduction Melanocortin-4 receptor (MC4R) gene is located on chromosome 18q22 [1]. MC4R, a G protein-coupled receptor is expressed in the developing autonomic and central nervous systems [2]. Activation of melanocortin-4- receptors (MC4Rs) reduces body fat stores by decreasing food intake and increasing energy expenditure [1]. Activation of MC4R proteins reduces body fat stores by decreasing food intake and increasing energy expenditure [1]. Mutations in the MC4R leads to a reduced receptor function found in 2–4% of extremely obese individuals [3]. Previous studies demonstrated several MC4R variants and common genetic polymorphisms near the MC4R gene contributing to different levels of obesity [4]. Recent genome wide scans found common variants near MC4R were related to obesity and insulin resistance such as rs17782313, rs17700633, rs12970134, rs477181, rs502933, and rs4450508. Among these variants, rs12970134 (A/G) located 154 kb downstream of MC4R has been studied most often. Many studies reported the association of rs12970134 MC4R variant with several obesity-related traits (such as: waist circumference, BMI) [4], [5], central obesity [6], [7] and insulin resistance [4], polycystic ovary syndrome [8], coronary artery disease [9]. Whe ... otide at the 3’-end of the primer perfectly complements the base at the variant or wild-type sequences. After the PCR and electrophoresis, the patterns of specific PCR products allow the differentiations of the SNP to determine whether the genotype was homozygous wild type, heterozygous or homozygous variant [14]. This method is relatively cheaper than other available methods. However, in order to create a working AS-PCR-based genotyping system, it needs to design primers and well optimized PCR conditions. Thus, this study aimed to find out a protocol for genotyping rs12970134 near MC4R by using AS-PCR and analyze the genotype and allele ratio of this SNP in Hanoi preschool chidren population. 2. Research methodology 2.1. Subjects and data collection In this study, 300 preschool children (36-60 months of age, 150 males and 150 females) in Hanoi with normal nutritional status, randomly were selected from the subjects of project B2018-SPH50 - a cross-sectional study identifying Kinh ethnic representing the major ethnic of Vietnam. Classification of nutrition status of children was performed according to WHO 2006 criteria; normal nutritional status were children with Z-score BMI by age and gender ranged from -2 to 2. Nguyen Thi Trung Thu et al. TNU Journal of Science and Technology 225(05): 38 - 44 Email: jst@tnu.edu.vn 40 The exclusion criteria were children with acute or chronic diseases such as tuberculosis, HIV/AIDS. Samples of cheek mucosa cells were taken with the consent of parents or official guardians. The project was approved by the Medical Ethics Council of the Institute of Nutrition with Decision No. 343/VDD-QLKH on July 27, 2018. 2.2. DNA extraction method DNA was extracted from the sample of the cheek mucosa cell by using the GeneJET Genomic DNA Purification kit (Thermo, USA) according to the manufacturer's instructions. 2.3. Genotyping method Protocol of genotyping SNP rs1137101 by AS- PCR method included the following steps: 2.3.1. Design primers Nucleotide sequence of DNA fragments containing SNP rs12970134 on NCBI database [15] was used to design three of PCR primers (including wildtype and mutant primers to detect 2 alleles (G or A) and one common primer. Designing wildtype and mutant primers to identify G or A allele of rs12970134 near MC4R gene was based on Wangkuhang et al. [16]. Next, last primer was designed by using Oligo 7 Primer Analysis Software [17] and UCSC In-Silico PCR online [18] to choose pairs of primers that have homologous melting temperature (Tm) and don’t match each other. The melting temperature of the primers was approximately 54°C according to recommendations of the above software. 2.3.2. Optimal protocol design for polymerase chain reaction To genotype rs12970134 polymorphism using AS-PCR, each genotype was determined by two independent reactions of A and G alleles. The composition of each PCR reaction consists of 0.8 µL of nuclease-free water, 2.5 µL master mix Dream Taq Green (containing: 0.4 mM Dream Taq DNA polymerase, 0.4 mM 2X Dream Taq Green buffer, 0.4 mM dATP, 0.4 mM dCTP, 0.4 mM dGTP, 0.4 mM dTTP and 4 mM MgCl2), 0.35 µL for each primer (concentration 10 pmol), 1.5 µL of DNA sample (concentration 37-60 ng/µl) in a total volume of 5.5 µL. The gradient PCR method was used to determine the annealing temperature. The PCR conditions were as follows: 3 min at 94 o C, 34 cycles of 30 sec at 94 0 C, 30 sec at 52 o C/54 o C/56 o C, 30 sec at 72 o C, final extension 8 min at 72 o C, chilling at 4 o C. PCR products (208-bp band) were detected on the redsafe-stained 2.5% agarose gel by the electrophoresis in 0.5X TBE buffer. The DNA band was taken by using Geldoc-It TM gel camera. The optimal protocol was recruited from the results of trials. 2.4. Statistical analysis Genotype and allele frequencies are expressed in %. Add the Hardy-Weinberg equation to identify allele frequencies. 3. Results and discussion 3.1. Optimize the protocol of genotyping MC4R rs12970134 3.1.1. Design the primers The results showed 5 oligos used to perform the AS-PCR. But all products were so short, that we only chose wildtype and mutant forward primers. The sequences of wildtype and forward primers were 5’- TCTTACCAAACAAAGCATGTG-3’, and 5’-TCTTACCAAACAAAGCATGTA-3’. The sequence of common reverse primer was 5’- GTCATTCCCACTACCACCTG-3’. The theorical PCR product was a 208-bp in length: TCTTACCAAACAAAGCATGT(G/A)caaac aaagatttatcagaagggtgcttgttagtacctgtattcaaaggg agaactagtcaaacctcaaaggggcaaggccaaccaggacc aacctagcagggcaagcatgtctccacactgcctatcttcagat gagcatttttttcctttaggcaagtttttcCAGGTGGTAG TGGGAATGAC Nguyen Thi Trung Thu et al. TNU Journal of Science and Technology 225(05): 38 - 44 Email: jst@tnu.edu.vn 41 3.1.2. Determination of the annealing temperature of the gradient PCR Figure 1. Electrophoresis image of gradient PCR products Ta: Temperature of annealing, (-): negative control, M: CSL-MDNA-100bp DNA Ladder RTU Genotype: 1 (GG), 2 (AG), 3 (AA) Figure 1 showed that at Ta = 54 o C, the amplified band was the thickest and easy to determine G and A alleles. Thus, the optimized PCR protocol was 94 o C for 3 min and 34 cycles of denaturation at 94 o C for 30 sec, primer annealing at 54 o C for 40 sec, primer extension at 72 o C for 30 sec, final extension at 72 o C for 8 min, stopped by chilling at 4 o C. 3.2. Result of validation To validate AS-PCR method, two products of A allele from sample 2 and G allele from sample 1 were verified by sequencing with reverse primer: 5’- GTCATTCCCACTACCACCTG-3’ (Figure 2). The obtained sequences from sequencing are single strands and complementary to the single PCR product sequence above (3.1.1). Genotypes were analyzed by AS-PCR method Result of sequencing method Allele A (sample 2) Allele G (Sample 1) Figure 2. Allele A and G products were validated by sequencing method Thus, the genotypes identified by using AS-PCR method were completed in concordance with those of the sequencing method. Consequently, we used the optimized AS-PCR protocol to genotype 200 samples and the results are presented in Figure 3. Nguyen Thi Trung Thu et al. TNU Journal of Science and Technology 225(05): 38 - 44 Email: jst@tnu.edu.vn 42 Figure 3. Electrophoresis image of AS-PCR products with some samples (-) : negative control, M: CSL-MDNA-100bp DNA Ladder RTU Genotype: 1 (GG), 2 (AG), 3 (AA), 4 (AG), 5 (GG), 6 (AG), 7 (GG), 8 (GG), 9 (GG), 10 (GG), 11 (GG), 12 (GG), 13 (GG), 14 (GG), 15 (GG), 16 (GG), 17 (AG), 18 (GG). 3.3. Frequencies of MC4R rs12970134 polymorphism in Hanoi preschool children Children were chosen equally by gender (50% boy) and age group (3-5 years: 25% (3–3.5 years), 25% (3.5–4 years), 25% (4–4.5 years), 25% (4.5–5 years)). The anthropometric indicators of 200 normal children (WHO 2006 criteria) were represented in our current publication [19]. The frequencies of genotypes and alleles of MC4R rs12970134 polymorphism among these children is shown in Table 1. Table 1. Genotype and allele frequencies of MC4R rs12970134 polymorphism in Hanoi 3-5 years old children MC4R rs12970134 Number (Frequencies) Genotype GG 115 (57.5%) AG 77 (38.5%) AA 8 (4.0%) Allele G 307 (77%) A 93 (23%) P HWE 0.265 The data in the table are presented by n (%), HWE: Hardy-Weinberg equation, P value were from test 2 or Fisher exact. In the entire samples, the GG genotype was the highest (57.5%), and the AA genotype was the lowest (4%). The frequencies of the G and A alleles were 0.77 and 0.23, respectively. The frequencies of rs12970134 genotypes in samples of preschool children in Hanoi were in the Hardy – Weinberg distribution (P = 0.265). The frequencies of rs12970134 genotypes in different populations varied significantly around the world [20]. The heterogeneity of the proportions of alleles in different populations was influenced by ethnic characteristics. According to Marth (2004), the history and characteristics of nation formation have a great influence on its biological characteristics, anthropology, and genetic background [21]. In the Hanoi primary school children population, the frequency of minor A allele was 0.23. This result was the same with other populations [20]. The ratio of genotypes in our study is almost equal to the frequencies in the Japanese in Tokyo, Japan (JPT) [20]. The limitation of this study is that the genotyping method identifies only one SNP genotype for one procedure. Also in this study, the frequencies of alleles and genotypes were determined only in children with normal physical development in Hanoi, and not the entire population of Vietnam. Further research is required large samples that are representative of the entire population, and the inclusion of children with nutritional disorders, children from different ethnic groups. Nguyen Thi Trung Thu et al. TNU Journal of Science and Technology 225(05): 38 - 44 Email: jst@tnu.edu.vn 43 4. Conclusions This research shows the AS-PCR method for genotyping MC4R rs12970134 polymorphism in Vietnam’s laboratories. This protocol used the forward primers (5’- tcttaccaaacaaagcatgtg-3’ to detect allele G, and 5’-tcttaccaaacaaagcatgta-3’ to detect allele A), and the reverse primer (5’- gtcattcccactaccacctg-3’). The temperature to anneal primer was 54 0 C. Among children aged 3-5 years in Hanoi, the frequency of GG genotype was the highest (57.5%) and the frequency of AA genotype was the smallest (4%). The method for identifying genotypes in this study was developed and optimized to ensure data accuracy, reduce costs, and can be used in many molecular biology laboratories to identify the MC4R rs12970134 genotype with large sample sizes. Acknowledgments The authors would like to thank Laboratory Center at Institute for Preventive Medicine and Public Health for kindly helps and supports. The study was supported by Ministry of Education and Training, Vietnam, grant no B2018 - SPH50. The research protocol was approved by The Ethics Committee of the National Institute of Nutrition, Vietnam. REFERENCES [1]. N. Balthasar et al., "Divergence of melanocortin pathways in the control of food intake and energy expenditure," Cell, vol. 123, no. 3, pp. 493-505, 2005. [2]. K. G. Mountjoy and J. M. Wild, "Melanocortin-4 receptor mRNA expression in the developing autonomic and central nervous systems," Developmental brain research, vol. 107, no. 2, pp. 309-314, 1998. [3]. F. Geller et al., "Melanocortin-4 receptor gene variant I103 is negatively associated with obesity," The American Journal of Human Genetics, vol. 74, no. 3, pp. 572-581, 2004. [4]. J. C. Chambers et al., "Common genetic variation near MC4R is associated with waist circumference and insulin resistance," Nature genetics, vol. 40, no. 6, p. 716, 2008. [5]. D. Albuquerque, C. Nóbrega, R. Rodríguez- López, and L. Manco, "Association study of common polymorphisms in MSRA, TFAP2B, MC4R, NRXN3, PPARGC1A, T MEM18, SEC16B, HOXB5 and OLFM4 genes with obesity-related traits among Portuguese children," Journal of human genetics, vol. 59, no. 6, p. 307, 2014. [6]. L. F. Been et al., "Replication of association between a common variant near melanocortin‐4 receptor gene and obesity‐related traits in Asian Sikhs," Obesity, vol. 18, no. 2, pp. 425-429, 2010. [7]. O. P. Dwivedi et al., "Strong influence of variants near MC4R on adiposity in children and adults: a cross-sectional study in Indian population," Journal of human genetics, vol. 58, no. 1, pp. 27, 2013. [8]. A. A. Batarfi et al., "MC4R variants rs12970134 and rs17782313 are associated with obese polycystic ovary syndrome patients in the Western region of Saudi Arabia," BMC medical genetics, vol. 20, no. 1, pp. 144, 2019. [9]. H. Huang et al., "Implication of genetic variants near TMEM18, BCDIN3D/FAIM2, and MC4R with coronary artery disease and obesity in Chinese: a angiography-based study," Molecular biology reports, vol. 39, no. 2, pp. 1739-1744, 2012. [10]. M. Bazzi et al., "Association between FTO, MC4R, SLC30A8, and KCNQ1 gene variants and type 2 diabetes in Saudi population," Genet Mol Res, vol. 13, no. 4, pp. 10194- 10203, 2014. [11]. B. Xi et al., "Association between common polymorphism near the MC4R gene and obesity risk: a systematic review and meta- analysis," PloS one, vol. 7, no. 9, p. e45731, 2012. [12]. D. P. Zobel et al., "Variants near MC4R are associated with obesity and influence obesity- related quantitative traits in a population of middle-aged people: studies of 14,940 Danes," Diabetes, vol. 58, no. 3, pp. 757-764, 2009. [13]. R. J. Loos et al., "Common variants near MC4R are associated with fat mass, weight and risk of obesity," Nature genetics, vol. 40, no. 6, p. 768, 2008. [14]. M. N. Darawi et al., "Allele-specific polymerase chain reaction for the detection of Alzheimer’s disease-related single nucleotide Nguyen Thi Trung Thu et al. TNU Journal of Science and Technology 225(05): 38 - 44 Email: jst@tnu.edu.vn 44 polymorphisms," BMC medical genetics, vol. 14, no. 1, p. 27, 2013. [15]. National Center for Biotechnology Information, "dbSNP short genetic variations", 2019. [Online]. Available: https://www.ncbi.nlm.nih.gov/projects/SNP/s np_ref.cgi?do_not_redirect&rs=rs12970134. [Accessed Aug. 11, 2019]. [16]. P. Wangkumhang et al., "WASP: a Web- based Allele-Specific PCR assay designing tool for detecting SNPs and mutations," BMC Genomics, vol. 8, no. 1, pp. 275-283, 2007. [17]. W. Rychlik, Oligo 7. [CD-ROM]. Cascade, Co: Molecular Biology Insights, 2009. [18]. J. Kent, "UCSC In-Silico PCR", 2013. [Online]. Available: d=start. [Accessed Aug. 12, 2019]. [19]. D. T. T. Le, T. T. T. Nguyen, N. V. Savvina, and T. T. Le, "Genotyping method and frequency of genotypes of single nucleotide polymorphism of the leptin receptor gene (LEPR-rs1137101) in preschool children in Vietnam," (In Russia), Pediatria named after G.N. Speransky, vol. 99, no. 1, pp. 121-126, 2020. [20]. MediaWiki and National Center for Biotechnology Information, “SNPedia”, Dec. 07, 2019. [Online]. Available: https://www.snpedia.com/index.php/Rs12970 134. [Accessed Feb. 7, 2020]. [21]. G. T. Marth, E. Czabarka, J. Murvai, and S. T. Sherry, "The allele frequency spectrum in genome-wide human variation data reveals signals of differential demographic history in three large world populations," Genetics, vol. 166, no. 1, pp. 351-372, 2004.
File đính kèm:
 genotyping_method_and_frequency_of_single_nucleotide_polymor.pdf
genotyping_method_and_frequency_of_single_nucleotide_polymor.pdf

